There will always be men destined to be subjugated by the opinions of their century, of their Country, of their Society: Some men today act the part of the Freethinker and the Philosopher who, for the same reason, would have been but fanatics at the time of the League.
--Rousseau, Preface to First Discourse
Computational modeler and miniature golf champion Eric-Jan Wagenmakers recently penned a short essay on a recent series of unfortunate events which have left social psychology with its fair share of bruises and black eyes. While social psychologists may tell you that they just fell down the stairs, the real cause of these contusions, says Wagenmakers, is more systemic.
First, flagship social psychology journals such as JPSP refuse to publish replication studies which can refute existing studies - an important scientific correction mechanism if there ever was one. This has led to the continued credibility of controversial studies such as Daryl Bem's famous (infamous?) precognition experiment, with studies reporting a failure to replicate these results being summarily rejected; apparently, unless a study can add something besides a failure to replicate, it is deemed unfit for publication. It would seem that providing evidence against an established theory would be valuable enough in and of itself, but the guidelines of some of the major journals say otherwise.
Second, there was the high-profile case of Diederik Stapel, a social psychology researcher who may have fabricated data affecting over thirty experiments. Cases of massive fraud are not specific to social psychology by any means - take Marc Hauser in evolutionary biology, for example - but it certainly doesn't help, especially in a field dealing with sensitive social and political topics which are likely to receive more attention from the public.
Third, eminent social psychologist John Bargh published a not-so-nice critique on a group of researchers who failed to replicate one of his experiments, as well as the journal which published it (PLoS ONE) and a commentator who covered the story. (A post about the event can be found here; it looks as though the original post by Bargh was deleted.) The ad hominem attacks employed by Bargh serve as an example of what not to do when someone fails to replicate your study; after all, replication is one of the pillars of scientific investigation. We're dealing with falsifiable hypotheses here, not instant Gospel every time a big name publishes a result. (I would, however, try to make an argument about how Bargh couldn't help writing that response due to uncontrollable external forces.)
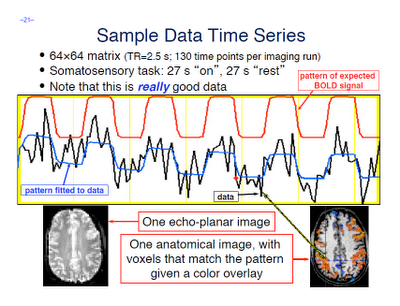
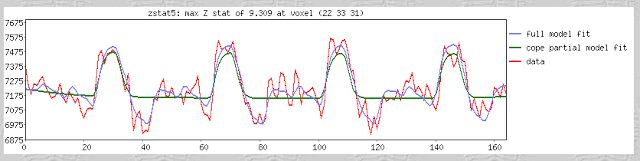
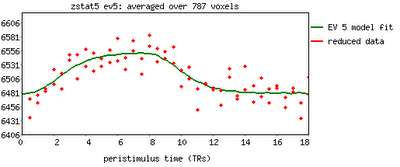
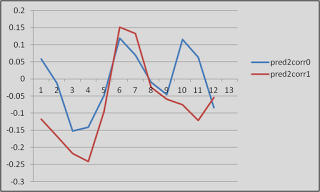
Fourth, scientists in the social and cognitive neuroscience fields are very good at obtaining the result that they want, whether they are aware of it or not. According to Wagenmakers: "...If you set out to torture the data until they confess, you will more likely than not obtain some sort of confession – even if the data are perfectly innocent." This is absolutely true, at least in my own experience. Usually I don't even realize when I am doing it. For example, I'll look at a few dozen different uncorrected voxel thresholds, in order to get it just right so that those two clusters are just touching so that they can pass a certain correction threshold; until I realize that, baby, those aren't activation blobs on my computer screen I'm looking at - it's specks of Cheetos dust. (Don't tell me this hasn't happened to you!)
Thus, the obvious corrections to all of the above are: 1) Journals should be more accepting of replication studies, in order to provide more incentive to replicate and also to mitigate the file-drawer problem; 2) Don't commit massive fraud and ruin the careers of your students and colleagues (but if you do, don't get caught); 3) If someone can't replicate your results, keep it civil; and 4) Make the switch to Wheat Thins. No, wait! What I meant was, try to set a standard of what you will do for your analysis, and adhere to that analysis plan. Wagenmakers recommends trying pre-registration of your experiment online, similar to what is being done with clinical work at the NIH; you may be surprised at how your analysis turns out. Whether the surprise will be good or bad is another matter.
Anyway, I recommend giving the original article a read; what you've just read is an inferior version of his prose. Also, Wagenmakers and Michael Lee have posted a free pdf of how to do Bayesian data analysis; I recommend taking a look at this as well, in case you are curious.
--Rousseau, Preface to First Discourse
Computational modeler and miniature golf champion Eric-Jan Wagenmakers recently penned a short essay on a recent series of unfortunate events which have left social psychology with its fair share of bruises and black eyes. While social psychologists may tell you that they just fell down the stairs, the real cause of these contusions, says Wagenmakers, is more systemic.
First, flagship social psychology journals such as JPSP refuse to publish replication studies which can refute existing studies - an important scientific correction mechanism if there ever was one. This has led to the continued credibility of controversial studies such as Daryl Bem's famous (infamous?) precognition experiment, with studies reporting a failure to replicate these results being summarily rejected; apparently, unless a study can add something besides a failure to replicate, it is deemed unfit for publication. It would seem that providing evidence against an established theory would be valuable enough in and of itself, but the guidelines of some of the major journals say otherwise.
Second, there was the high-profile case of Diederik Stapel, a social psychology researcher who may have fabricated data affecting over thirty experiments. Cases of massive fraud are not specific to social psychology by any means - take Marc Hauser in evolutionary biology, for example - but it certainly doesn't help, especially in a field dealing with sensitive social and political topics which are likely to receive more attention from the public.
Third, eminent social psychologist John Bargh published a not-so-nice critique on a group of researchers who failed to replicate one of his experiments, as well as the journal which published it (PLoS ONE) and a commentator who covered the story. (A post about the event can be found here; it looks as though the original post by Bargh was deleted.) The ad hominem attacks employed by Bargh serve as an example of what not to do when someone fails to replicate your study; after all, replication is one of the pillars of scientific investigation. We're dealing with falsifiable hypotheses here, not instant Gospel every time a big name publishes a result. (I would, however, try to make an argument about how Bargh couldn't help writing that response due to uncontrollable external forces.)
Fourth, scientists in the social and cognitive neuroscience fields are very good at obtaining the result that they want, whether they are aware of it or not. According to Wagenmakers: "...If you set out to torture the data until they confess, you will more likely than not obtain some sort of confession – even if the data are perfectly innocent." This is absolutely true, at least in my own experience. Usually I don't even realize when I am doing it. For example, I'll look at a few dozen different uncorrected voxel thresholds, in order to get it just right so that those two clusters are just touching so that they can pass a certain correction threshold; until I realize that, baby, those aren't activation blobs on my computer screen I'm looking at - it's specks of Cheetos dust. (Don't tell me this hasn't happened to you!)
Thus, the obvious corrections to all of the above are: 1) Journals should be more accepting of replication studies, in order to provide more incentive to replicate and also to mitigate the file-drawer problem; 2) Don't commit massive fraud and ruin the careers of your students and colleagues (but if you do, don't get caught); 3) If someone can't replicate your results, keep it civil; and 4) Make the switch to Wheat Thins. No, wait! What I meant was, try to set a standard of what you will do for your analysis, and adhere to that analysis plan. Wagenmakers recommends trying pre-registration of your experiment online, similar to what is being done with clinical work at the NIH; you may be surprised at how your analysis turns out. Whether the surprise will be good or bad is another matter.
Anyway, I recommend giving the original article a read; what you've just read is an inferior version of his prose. Also, Wagenmakers and Michael Lee have posted a free pdf of how to do Bayesian data analysis; I recommend taking a look at this as well, in case you are curious.